Converting Fat to Fight Obesity
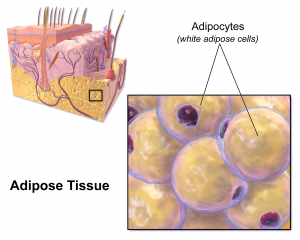
There are two types of fat in the human body: brown and white. Brown fat, the “good” fat, is rich in mitochondria, which gives it its brown appearance. Whereas white fat stores calories and acts as an insulator, mitochondria-rich brown fat burns energy to produce heat throughout the body and maintains body temperature. White fat, conversely, uses its stored energy to insulate the body and keep its temperature level. While all fat serves a purpose in the body, an excess of white fat cells causes obesity, a condition affecting one in three adults in the U.S. and the root cause of many potential health problems. Finding ways to convert white fat to brown opens a possibility of treating this problem naturally.
A new study in Scientific Reports proposes a clever way to convert fat types. Professor of Biomedical Engineering Samuel Sia, PhD, of the Columbia University School of Engineering and Applied Science, led a team which developed a method of converting white fat into brown using a tissue-grafting technique. After extracting and converting the fat, it can then be transplanted back into the patient. White fat is hard-wired to convert to brown under certain conditions, such as exposure to cold temperatures, so the trick for Dr. Sia’s team was finding a way to make the conversion last for long periods. The studies conducted with mice suggested that using these methods, newly-converted fat stayed brown for a period of two months.
Dr. Sia’s team will proceed to conduct further tests, especially on the subjects’ metabolism and overall weight after undergoing the procedure, and they hope that eventual clinical trials will result in new methods to treat or even prevent obesity in humans.
Cremins Lab Student Appointed Blavatnik Fellow

The Perelman School of Medicine named Linda Zhou, a student in BE’s Cremins Laboratory, a Blavatnik Fellow for the 2018-2019 academic year. The selection process for this award is highly competitive, and Linda’s selection speaks to the excellent quality of her scholarship and academic performance. The fellows will be honored in a special ceremony at the Museum of Natural History in New York City.
Linda received her B.S. in Biophysics and Biochemistry from Yale University and is currently pursuing her M.D./Ph.D. in the Genomics and Computational Biology Program at Penn. “I am honored to be named a Blavatnik Fellow and am extremely excited to continue my graduate studies investigating neurological disorders and the 3D genome,” she said. “This support will be integral to achieving my long term goal of driving scientific discovery that will help treat human disease.”
Linda’s research is overseen by Penn Bioengineering Assistant Professor Jennifer Phillips-Cremins, PhD. “Linda is an outstanding graduate student,” said Dr. Cremins. “It is a true delight to work with her. She is hard working, intelligent, kind, and has extraordinary leadership ability. Her unrelenting search for ground-state truth makes her a shining star.”
The Blavatnik Family Fellowship in Biomedical Research is a new award announced by the Perelman School of Medicine in May of this year. This generous gift from the Blavatnik Family Foundation awards $2 million to six recipients in the Biomedical Graduate Studies Program at Penn for each of the next four years.
Growing Lungs in a Lab
As the demand for lung transplants continues to rise, so does the need for safe and effective transplanted lungs. Bioengineered lungs grown or created in labs are one way of meeting this demand. The problem – as is ever the case with transplants – is the high rate of rejection. The results of success are always better when cells from the patient herself (or autologous cells) are used in the transplanted organ.
Recently Joan Nichols, PhD, Professor of Internal Medicine, and Microbiology and Immunology, at the University of Texas Medical Branch at Galveston, successfully bioengineered the first human lung. Her latest study published in Science Translational Medicine describes the next milestone for Dr. Nichols’ lab: successfully transplanting a bioengineered lung into a pig.
These advances are possible due to Dr. Nichols’ work with autologous cells, continuing the trend of “on demand” medicine (i.e. medicine tailor for a specific patient) which we track on this blog. Dr. Nichols’ particular method is to build the structure of a lung (using the harvested organs of dead pigs in this case), de-cellularize the tissue, and then repopulate it with autologous cells from the intended recipient. This way, the host body recognizes the cells as friendly and the likelihood of acceptance increases. While further study is needed before clinical trials can begin, Dr. Nichols and her team see the results as extremely promising and believe that we are on the way to bioengineered human lungs.
Nanoparticles Combat Dental Plaque
Combine a diet high in sugar with poor oral hygiene habits and dental cavities likely result. The sugar triggers the formation of an acidic biofilm (plaque) on the teeth, eroding the surface. Early childhood dental cavities affect one in every four children in the United States and hundreds of millions more globally. It’s a particularly severe problem in underprivileged populations.
In a study published in Nature Communications this week, researchers led by Hyun (Michel) Koo of the University of Pennsylvania School of Dental Medicine in collaboration with David Cormode of Penn’s Perelman School of Medicine and School of Engineering and Applied Science used FDA-approved nanoparticles to effectively disrupt biofilms and prevent tooth decay in both an experimental human-plaque-like biofilm and in an animal model that mimics early-childhood caries.
Dr. David Cormode is Assistant Professor of Radiology and Secondary Faculty in Bioengineering at Penn. His research includes Bioengineering Therapeutics, Devices and Drug Delivery and Biomaterials.
Read the full story at Penn Today. Media contact Katherine Unger Baillie.
Stopping the Flu from Catching On
The flu virus is notoriously contagious, but there may be a way to stop it before it starts. In order for the influenza virus to successfully transport itself into the cells of a human host, it needs a certain protein called hemagglutinin which mediates its entry. By interfering with this vital ingredient, researchers can effectively kill the virus.
A new study in the Proceedings of the National Academy of Sciences discusses a method of disrupting the process by which this protein causes the virus to infect its host cells. This discovery could lead to more effective flu vaccines that target the flu virus at its root, rather than current ones which have to keep up with the ongoing changes and mutations of the virus itself. Indeed, the need for different vaccines to address various “strains” of the flu is moot if a vaccine can stop the virus from infecting people in the first place.
This breakthrough results from grants provided by the NSF, the Welch Foundation, and the NIH to Rice University and Baylor College of Medicine. Lead researchers José Onuchic, PhD, Harry C. and Olga K. Wiess Chair of Physics and Professor of Chemistry and BioSciences at Rice University; Jianpeng Ma, PhD, Professor of Bioengineering at Rice University and Lodwick T. Bolin Professor of Biochemistry at Baylor College of Medicine; and Qinghua Wang, PhD, Assistant Professor of Biochemistry at Baylor College of Medicine. Their team will continue to study the important role proteins play in how the flu virus operates.
People and Places
This week, we congratulate a few new leadership appointments in bioengineering. First, the Georgia Institute of Technology appointed Penn BE alumnus Andréas García, PhD, the new Executive Director of the Parker H. Petit Institute for Bioengineering and Bioscience. In addition to his new role, Dr. García is also the George W. Woodruff School of Mechanical Engineering Regents Professor. He conducts research in biomolecular, cellular, and tissue engineering and collaborates with a number of research centers across Georgia Tech. Dr. García graduated with both his M.S.E. and Ph.D. from the University of Pennsylvania’s Department of Bioengineering.
Secondly, the University of Minnesota Institute for Engineering in Medicine (IEM) named the Distinguished McKnight University Professor John Bischof, PhD, their new director. This follows Dr. Bischof’s recent position as interim director for the IEM. Dr. Bischof earned his Ph.D. in Mechanical Engineering at the University of California at Berkeley, and is currently a faculty member in both the Mechanical Engineering and Biomedical Engineering Departments at the University of Minnesota. Dr. Bischof holds the Carl and Janet Kuhrmeyer Chair in Mechanical Engineering.
At an earlier, but no less impressive, point in his academic career, Tanishq Abraham became the youngest person to graduate with a degree in biomedical engineering. The fifteen year old recently graduated summa cum laude from the University of California, Davis. As part of his graduating research, Abraham – a first-generation Indian-American – designed a device to measure the heart rates of burn victims. Abraham has already been accepted by U.C. Davis for his Ph.D. and plans to continue on to his M.D.
Finally, the work continues to create affordable and well-fitted prosthetics, especially for remote, rural, and underfunded areas both in the U.S. and abroad. Unfortunately, recent studies published by the Centre for Biomedical Engineering at the India Institute of Technology Delhi (IIT) demonstrate the uphill nature of this battle; stating that India alone contains over half a million upper limb amputees. To address this explosive population, researchers and entrepreneurs are using new bioengineering technologies such as digital manufacturing, 3D scanning and printing, and more. The best innovations are those that save time, resources, and money, without sacrificing quality in the prosthetic or patient comfort. Penn Engineering’s Global Biomedical Service (GBS) program similarly responds to this need, as each year students follow an academically rigorous course with a two-week immersive trip to China, where they learn how to create and fit prosthetic limbs for local children in conjunction with Hong Kong Polytechnic University.



 As new technologies emerge, whether related to health care, artificial intelligence, or other aspects of society, they bring with them new ethical challenges.
As new technologies emerge, whether related to health care, artificial intelligence, or other aspects of society, they bring with them new ethical challenges.




