Pain may be a universal experience, but what actually causes that experience within our brains is still poorly understood. Pain often continues long after the relevant receptors in the body have stopped being stimulated and can persist even after those receptors cease to exist, as is the case with “phantom limb” pain.
The exact experience an individual will have after a painful incident comes down to the complex, variable connections formed between several different parts of the brain. The inability to predict how those connections will form and evolve can make pain management a tricky, frustrating endeavor for both healthcare providers and patients.
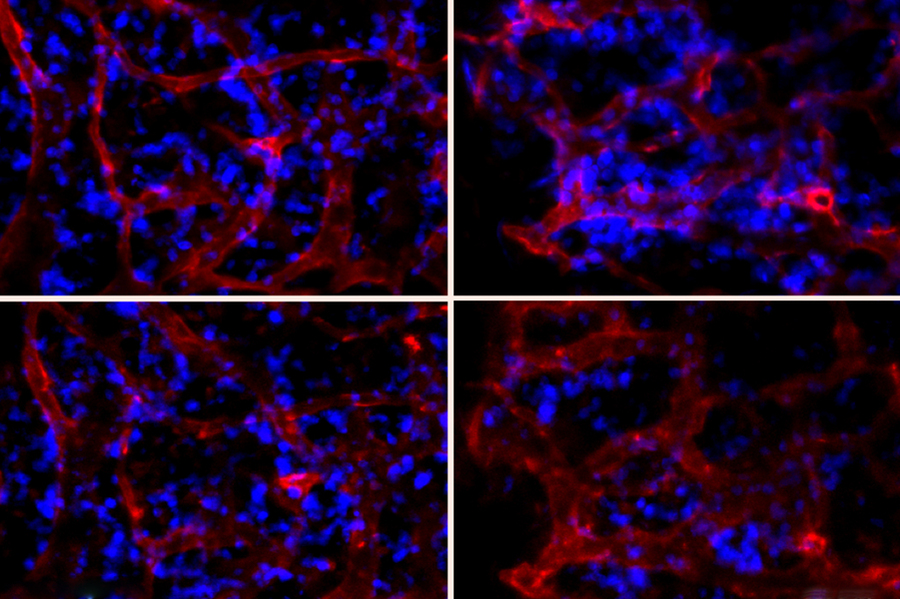
Now, a team of Penn researchers has shown a way to make such predictions from the pattern of neural connections that begin to take shape soon after the first onset of pain. Though their study was conducted in rats, it suggests that similar brain imaging techniques could be used to guide treatment decisions in humans, such as which individuals are most likely to benefit from different drugs or therapies.
The study, published in the journal Pain, was led by Beth Winkelstein, Eduardo D. Glandt President’s Distinguished Professor in Penn Engineering’s Department of Bioengineering and Deputy Provost of the University of Pennsylvania, along with Megan Sperry, then a graduate student in her lab. Eric Granquist, Director of the Center for Temporomandibular Joint Disease at the Hospital of the University of Pennsylvania in the Department of Oral & Maxillofacial Surgery, and assistant professor of Oral & Maxillofacial Surgery in Penn’s School of Dental Medicine, also contributed to the research.
“Our findings provide the first evidence that brain networks differ between acute and persistent pain states, even before those different groups of rats actually show different pain symptoms,” says Winkelstein.
Read the full story at Penn Engineering Today. Media contact Evan Lerner.