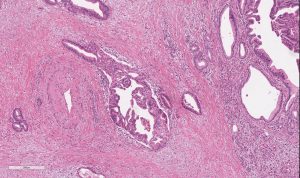
Swept up in a pancreatic cancer diagnosis is inevitably a sense of fear and sadness.
But at Penn, researchers are bringing new hope to this disease. And with patients like Nick Pifani, it’s clear that they’re moving in the right direction.
Pifani, from Delran, New Jersey, first noticed some lingering stomach upset in February 2017. He called his family doctor, concerned—especially given that he was an otherwise healthy marathon runner who was only 42. He was sent to a gastrointestinal specialist. A few weeks later, some crippling stomach pain sent him back to the emergency room and he received an MRI that showed a mass on his pancreas—Stage Three, inoperable, he was told.
He was treated with chemotherapy, along with radiation and, eventually, and after receiving advice from doctors at Penn, his tumor was removed. Thereafter, he realized he had a PALB2 mutation—a cousin of the BRCA gene mutation. At that moment, his long-term needs changed and he found himself seeking specialized care at Penn, where he met Kim Reiss Binder, assistant professor of medicine at the Hospital of the University of Pennsylvania (HUP).
“I’m a planner; I want to understand what [my] potential options are,” Pifani says. “[Reiss Binder] asked why I was there to see her and I explained and quickly I could tell she was—outside of her being remarkably intelligent—a great listener and a compassionate doctor.”
“I have a feeling she worries about me more than I do,” he laughs.
Pifani has now been in remission for two years and four months; he sees Reiss-Binder every three months for checkups. His survival story is inspiring and a sign of momentum, even if a world without pancreatic cancer is still frustratingly out of reach.
Pancreatic cancer at Penn
Pancreatic cancer is the third-leading cause of cancer-related death in the United States, outmatched only by lung cancer (No. 1) and colorectal cancer (No. 2). A person diagnosed with pancreatic cancer is still unlikely to survive past five years—only 9% of survivors do, giving it the highest mortality rate among every major cancer.
In short, pancreatic cancer seldom paves the way for optimistic narratives. Some of the hope that has surfaced, though, is thanks to some talent, dedication to the cause, and hard work at Penn.
A key point of progress in the battle against the disease was made in 2002, when former Assistant Professor of Medicine David Tuveson established a standard model for examining human development of this disease in mice. This model has allowed for a reliable way to study the disease and has influenced progress made here at Penn and elsewhere since.
“There’s been a burst of activity in translational research, from bench to bedside,” explains Ben Stanger, the Hanna Wise Professor in cancer research and director of the Penn Pancreatic Cancer Research Center (PCRC) at the Abramson Cancer Center.
“And there’s a lot of momentum with community building, a dramatic increase in patient volumes, and a dramatic increase in what we know about the cancer,” he says of the status of pancreatic cancer today.
Reiss Binder, meanwhile, explains that one mark of progress at Penn and beyond has been learning about people like Pifani, who have the PALB2 gene, and why they respond differently to treatments than those without it. Platinum-based chemotherapies, for example, are especially effective for people with the PALB2 gene who are battling pancreatic cancer. An ongoing trial at Penn has tested and found some success with using PARP inhibitors—taken orally as an enzyme that fixes single-stranded breaks of DNA—as a maintenance therapy in that same PALB2 demographic after they’ve had chemotherapy. These are less toxic than chemotherapy for patients with the same mutations.
It’s all been slow progress toward better treatments, but there has been progress.
“This is the tip of the iceberg for a disease that we historically have treated with perpetual chemotherapy,” Reiss Binder says. “We owe it to patients to find better options to suppress the cancer but not ruin their quality of life.”
Catching cancer earlier
The consensus on why pancreatic cancer is so deadly? It just can’t be spotted fast enough.
Pancreatic cancer often presents well after it has developed and metastasized, and does so in a way that is not easy to recognize as cancer. Common symptoms include, for example, stomach upset and back pain. And by the time a harder-to-ignore symptom of the cancer surfaces, a sort of yellowing of the skin (a result of a bile duct blockage), it’s likely too late to stop the cancer in its tracks.
One approach to improved detection being tested at Penn, by Research Assistant Professor of Medicine Erica Carpenter, is a liquid biopsy—drawn from a standard blood test. Current means to test for pancreatic cancer—imaging through an endoscopic tube—are invasive and expensive, meaning a common liquid test could transform how many cases are detected early.
Carpenter explains that circulating tumor cells (CTCs) can shed from a tumor that’s adjacent to the wall of a blood vessel; what’s shed then shows up in a blood test. The cells, if detected, can explain more about the nature of the tumor, giving doctors an opportunity to examine characteristics of cancerous cells and decide how to effectively treat a tumor if it can’t be surgically removed. It also allows interpretations of disease burden and the effectiveness of medications—through genome sequencing—that imaging does not.
Ultimately, this gives doctors the potential to track the growth of a tumor before it’s fully developed, all through one tube of blood—detected through an innovative use of technology.

David Issadore, associate professor of bioengineering and electrical and systems engineering in the School of Engineering and Applied Science, has worked since 2017 to develop a chip that detects cancer in the blood, using machine learning to sort through literally hundreds of billions of vesicles and cells, looking for these CTCs. The chip retrieves data and the machine learning developed interprets that data, attempting to make a diagnosis that not only finds pancreatic cancer but also provides information about its progression—and, importantly, whether a patient might benefit from surgery.
Read the full story at Penn Today. Media contact Brandon Baker.

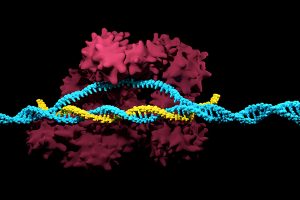

 A new technique that researchers from the Broad Institute of MIT and Harvard University are calling DNA microscopy could help map cells for better understanding of genetic and molecular complexities.
A new technique that researchers from the Broad Institute of MIT and Harvard University are calling DNA microscopy could help map cells for better understanding of genetic and molecular complexities. 

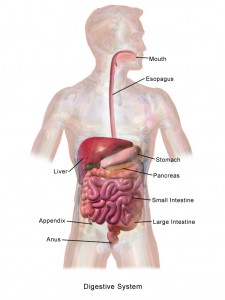 One of the critical organ transplant shortages in medicine is the gap between patients needing a liver transplant (around 13,000 each year) and the those receiving a transplant (about 7,000). For many years, bioengineers tried to build liver tissue in sophisticated 2D and 3D structures. Yet we never really knew how nature ‘interpreted’ these structures. A research team at Cincinnati Children’s Hospital led by Takanori Takebe, MD,
One of the critical organ transplant shortages in medicine is the gap between patients needing a liver transplant (around 13,000 each year) and the those receiving a transplant (about 7,000). For many years, bioengineers tried to build liver tissue in sophisticated 2D and 3D structures. Yet we never really knew how nature ‘interpreted’ these structures. A research team at Cincinnati Children’s Hospital led by Takanori Takebe, MD,