Using AI to Better Understand Cancer Immunology
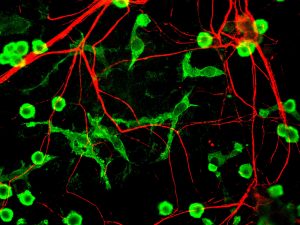
T lymphocytes in the immune system play a vital role in the body to recognize invasion by an outside element. When foreign bacteria enter the body, receptors on the T cell surface detect antigens associated with the bacteria and send a signal deploying phagocytes to attack and defeat the invading bacteria. While evolution and vaccination make the immune system very efficient, the inability of T cell receptors (TCRs) to detect cancer makes normal T cells relatively ineffective in resisting cancer. One of the ways to overcome this limitation of the immune system is to better understand how the TCRs respond to antigens. Analyses of the proteins involved in TSR responses are useful but limited by several factors, including the dizzying amount of data involved. Data analysis techniques have been helpful but have offered little information about the general reactions of TSRs, rather than how they react to specific antigens.
A possible solution to this obstacle is ImmunoMap, developed by scientists collaborating between Johns Hopkins University and Memorial Sloan Kettering Cancer Center. In a study recently published in Cancer Immunology Research, the authors, led by Jonathan P. Schneck, M.D., Ph.D., a professor of pathology at Johns Hopkins associated with the university’s Institute for Cell Engineering and Institute for Nanobiotechnology, describe their creation and deployment of ImmunoMap, a group of artificial intelligence algorithms that use machine learning to process large amounts of sequencing data and compare data from different antigens with each other.
The authors trained ImmunoMap initially using data from a mouse model of melanoma, in which the algorithm demonstrated significantly better performance than traditional methods. Subsequently, ImmunoMap was applied to patient response data from a melanoma clinical trial of the chemotherapy agent nivolumab. The algorithm discovered a new group of patients that would respond positively to nivolumab treatment — a finding missed by popular past methods. More testing of ImmunoMap is necessary, but the technology could make significant contributions to the monitoring of cancer patients receiving chemotherapy. In addition, it could to help to better predict response in patients before they begin specific chemotherapy regimes.
Wearables Improving Health
Among the most troubling health disparities related to global wealth inequality is the higher rate of mortality among children suffering from cancer. Fever is a common symptom of children undergoing cancer treatment, and this symptom may indicate more serious health issues that require the attention of a doctor. However, continuously monitoring skin temperature in children from low resource settings is difficult. Seeking to help remedy this problem, undergraduate engineering students at Harvard collaborated with the Dana-Farber/Boston Children’s Cancer & Blood Disorders Center’s Global Health Initiative to develop tools for earlier fever detection and treatment.
In a course taught by David Mooney, Ph.D., Robert P. Pinkas Family Professor of Bioengineering at Harvard, students developed a wearable device that sounds an alarm when the wearer needs medical help. The app can send patients’ recorded messages to their doctors, who can then review the temperature data and messages from the children before responding. Fashioned like a wristwatch, the extra-durable and waterproof device will next move into pilot testing among a larger patient population.
Meanwhile, at Northwestern, John A. Rogers, Ph.D., the Louis Simpson and Kimberly Querrey Professor of Materials Science and Engineering, Biomedical Engineering, and Neurological Surgery in Northwestern’s McCormick School of Engineering, has partnered with cosmetics giant L’Oréal to create the world’s smallest wearable. The device, which is smaller than an adult fingernail, measures UV sun exposure for the wearer and can tell when they should go back inside instead of risking overexposure. Unsurprisingly, it’s solar powered, and it was demonstrated a couple of weeks ago at a consumer electronics show in Las Vegas.
Growing Hydrogels Like Human Tissue
Scientists at Carnegie Mellon University and Nanyang Technological University in Singapore have collaborated in a process to create polyacrylamide gels that grow in a manner resembling natural tissue. K. Jimmy Hsia, Ph.D., Professor of Biomedical Engineering at Carnegie Mellon, is co-lead author of a new study in PNAS describing this new growth mode.
In the study, Dr. Hsia and his coauthors report that, in the same way that growth factors secreted by a living organism affect the generation of new tissue, oxygen can be modulated to control how hydrogels grow. Moreover, while growth is under way, the process could be continued to efficiently manage the mass transfer of nutrients from cell to cell. Finally, the authors detail the mechanical processes that help to shape the final product. With this new process, the ability to design and create materials for applications such as robotics and tissue engineering comes a step closer to resembling living tissue as closely as possible.
People and Places
Engineers at Virginia Tech have been awarded a $1.1 million grant from the Virginia Research Investment Committee to develop a device that uses low-energy electric fields for the treatment of brain tumors. Rafael Davalos, Ph.D., L. Preston Wade Professor of Biomedical Engineering and Mechanics, is the chief investigator on the grant.
The Department of Biomedical Engineering has announced the appointment of Kam W. Leong, Ph.D., as the Samuel Y. Sheng Professor of Biomedical Engineering. Dr. Leong earned his Ph.D. in chemical engineering from the University of Pennsylvania and taught at Duke and Johns Hopkins before arriving at Columbia in 2006. He was previously the James B. Duke Professor of Biomedical Engineering at Duke. Congratulations to Dr. Leong!