by Katherine Unger Baillie
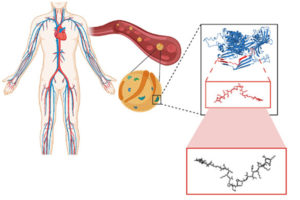
Fibrosis is the thickening of various tissues caused by the deposition of fibrillar extracellular matrix (ECM) in tissues and organs as part of the body’s wound healing response to various forms of damage. When accompanied by chronic inflammation, fibrosis can go into overdrive and produce excess scar tissue that can no longer be degraded. This process causes many diseases in multiple organs, including lung fibrosis induced by smoking or asbestos, liver fibrosis induced by alcohol abuse, and heart fibrosis often following heart attacks. Fibrosis can also occur in the bone marrow, the spongy tissue inside some bones that houses blood-producing hematopoietic stem cells (HSCs) and can lead to scarring and the disruption of normal functions.
Chronic blood cancers known as “myeloproliferative neoplasms” (MPNs) are one example, in which patients can develop fibrotic bone marrow, or myelofibrosis, that disrupts the normal production of blood cells. Monocytes, a type of white blood cell belonging to the group of myeloid cells, are overproduced from HSCs in neoplasms and contribute to the inflammation in the bone marrow environment, or niche. However, how the fibrotic bone marrow niche itself impacts the function of monocytes and inflammation in the bone marrow was unknown.
Now, a collaborative team from Penn, Harvard, the Dana-Farber Cancer Institute (DFCI), and Brigham and Women’s Hospital has created a programmable hydrogel-based in vitro model mimicking healthy and fibrotic human bone marrow. Combining this system with mouse in vivo models of myelofibrosis, the researchers demonstrated that monocytes decide whether to enter a pro-inflammatory state and go on to differentiate into inflammatory dendritic cells based on specific mechanical properties of the bone marrow niche with its densely packed ECM molecules. Importantly, the team found a drug that could tone down these pathological mechanical effects on monocytes, reducing their numbers as well as the numbers of inflammatory myeloid cells in mice with myelofibrosis. The findings are published in Nature Materials.
“We found that stiff and more elastic slow-relaxing artificial ECMs induced immature monocytes to differentiate into monocytes with a pro-inflammatory program strongly resembling that of monocytes in myelofibrosis patients, and the monocytes to differentiate further into inflammatory dendritic cells,” says co-first author Kyle Vining, who recently joined Penn’s School of Dental Medicine and School of Engineering and Applied Science as an assistant professor of preventive and restorative sciences. “More viscous fast-relaxing artificial ECMs suppressed this myelofibrosis-like effect on monocytes. This opened up the possibility of a mechanical checkpoint that could be disrupted in myelofibrotic bone marrow and also may be at play in other fibrotic diseases.”
Vining worked on the study as a postdoctoral fellow at Harvard in the lab of David Mooney. “Our study shows that the differentiation state of monocytes, which are key players in the immune system, is highly regulated by mechanical changes in the ECM they encounter,” says Mooney, who co-led the study with DFCI researcher Kai Wucherpfennig. “Specifically, the ECM’s viscoelasticity has been a historically under-appreciated aspect of its mechanical properties that we find correlates strongly between our in vitro and the in vivo models and human disease. It turns out that myelofibrosis is a mechano-related disease that could be treated by interfering with the mechanical signaling in bone marrow cells.”
Continue reading at Penn Today.