by Sophie Burkholder
 The George H. Stephenson Foundation Educational Laboratory and Bio-MakerSpace, more commonly known as the Bio-MakerSpace, has recently become a hub for Penn student start-ups that continue after graduation. Beyond offering a home base for projects by Bioengineering majors, the lab is also open to Penn students, regardless of major. Unlike other departmental undergraduate labs, the Bio-MakerSpace encourages interdisciplinary projects and collaborations from students across all different majors.
The George H. Stephenson Foundation Educational Laboratory and Bio-MakerSpace, more commonly known as the Bio-MakerSpace, has recently become a hub for Penn student start-ups that continue after graduation. Beyond offering a home base for projects by Bioengineering majors, the lab is also open to Penn students, regardless of major. Unlike other departmental undergraduate labs, the Bio-MakerSpace encourages interdisciplinary projects and collaborations from students across all different majors.
Even better, the lab has a neutral policy when it comes to intellectual property (IP), meaning all IP behind student projects belongs to the students instead of the lab or the engineering school. With a wide variety of prototyping equipment, coding and software programs installed on lab computers, and an extremely helpful lab staff, the Bio-MakerSpace provides students of all academic backgrounds the resources to turn their ideas into realities or even businesses, as a recent succession of start-ups founded in the lab has shown.
One of the most successful start-ups to come out of the Bio-MakerSpace in the last few years is Group K Diagnostics, founded by 2017 Bioengineering alumna Brianna Wronko. The company focuses on the use of a point-of-care diagnostic device called KromaHealthTM. Offering a variety of different tests based on the input of a small amount of blood, serum, or urine, the device induces a color change through a series of reactions that can be detected through image processing. Developed in part from Wronko’s senior design project (hence the name “Group K”) and in part from her experience working at an HIV clinic, Group K Diagnostics looks to expand access to care for all populations.
But not all start-ups from the Bio-MakerSpace have origins in senior design projects. Three start-ups from 2019, two of which won the Penn President’s Innovation Prize, all began as independent initiatives from students. InstaHub, founded by 2019 Wharton alumnus Michael Wong with help from Bioengineering doctoral candidate Dayo Adewole, is a company that focuses on the use of snap-on automation for light energy conservation. A simple and easy-to-install device with motion and occupancy sensors, InstaHub aims to reduce energy consumption in a way that’s simpler and cheaper than rewiring projects that might otherwise be required. Here, Adewole shares the way that access to the Bio-MakerSpace provided InstaHub with a helpful platform.
The second start-up from 2019 to come out of the Bio-MakerSpace and win a President’s Innovation Prize is Strella Biotechnology, founded by recent graduate Katherine Sizov (Biology 2019). In developing sensors with the ability to detect ethylene gas emitted by rotting fruits, Strella hopes to reduce the immense amount of food waste due to produce simply going bad in storage. With a patent-pending biosensor that mimics the way ripe fruits detect ethylene emissions of nearby rotting fruits, the technology behind Strella involves both biology and aspects of engineering. In this video, Sizov herself talks about the way that the Bio-MakerSpace opened its doors to her, and allowed her work to really take off with the help of resources she wouldn’t have easily found otherwise.
Yet another start-up to use the Bio-MakerSpace as a launch pad for innovation is BioAlert Technologies, comprised of a group of Penn engineering undergraduate and graduate students, including 2019 Bioengineering alumnus Johnny Forde and current Biotechnology student Marc Rosenberg, who is the startup’s CEO and founder. BioAlert’s innovations are in what they call continuous infection monitoring (CIM) systems, designed to detect infections in patients with diabetic foot ulcers. Often, even when properly bandaged by a doctor, these ulcers run the risk of bacterial infection once a patient returns home and continues to care for the wound. BioAlert uses their platform to assess whether or not a bacterial infection might occur in a given patient’s wound, and uses an app to alert both patients and doctors of it, so that patients can receive the proper response treatment and medication as quickly as possible.
Though each of these start-ups used the resources of the Bio-MakerSpace, they are each interdisciplinary approaches to solving real-world problems today. Paired with other student resources at Penn like courses offered under an Engineering Entrepreneurship minor, knowledge from the nearby Wharton business school professors, and competitions like the Rothberg Catalyzer, the Bio-MakerSpace allows for any student to transform their idea into a reality, and potentially take it to market.
Interested in learning more? Contact the BE Labs.



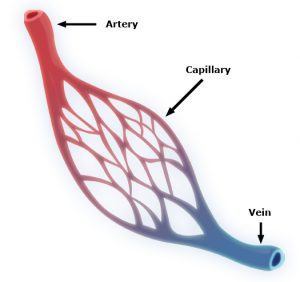 Capillaries are one of the most important forms of vasculature in our body, as they allow our blood to transfer nutrients to other parts of our body. But for how much effect capillary functionality can have on our health, their small size makes them extremely difficult to engineer into models for a variety of diseases. Now, researchers at the University of Washington led by
Capillaries are one of the most important forms of vasculature in our body, as they allow our blood to transfer nutrients to other parts of our body. But for how much effect capillary functionality can have on our health, their small size makes them extremely difficult to engineer into models for a variety of diseases. Now, researchers at the University of Washington led by 



 As technology and hands-on activities continue to become a larger part of education at all levels, a new movement of do-it-yourself projects is on the rise. Known as the “MakerSpace Movement,” the idea is that with the use of devices like 3-D printers, laser cutters, and simple circuitry materials, students, classes and communities can apply topics discussed in the classroom to real-life projects. Especially popular among STEM educators, the MakerSpace Movement is one that’s taken over labs in engineering schools around the country. Here at Penn, our own
As technology and hands-on activities continue to become a larger part of education at all levels, a new movement of do-it-yourself projects is on the rise. Known as the “MakerSpace Movement,” the idea is that with the use of devices like 3-D printers, laser cutters, and simple circuitry materials, students, classes and communities can apply topics discussed in the classroom to real-life projects. Especially popular among STEM educators, the MakerSpace Movement is one that’s taken over labs in engineering schools around the country. Here at Penn, our own  Believe it or not, however, some of the best parts of the Bio-MakerSpace can actually be purchased for a total of under $1500. Though that number is probably far beyond the individual budget of most students, it might be more affordable for a student club or dorm floor that receives additional funding from Penn. While the idea of building a MakerSpace from nothing might sound intimidating, the popularity of the movement actually helps to provide a wide range of technology and affordable options.
Believe it or not, however, some of the best parts of the Bio-MakerSpace can actually be purchased for a total of under $1500. Though that number is probably far beyond the individual budget of most students, it might be more affordable for a student club or dorm floor that receives additional funding from Penn. While the idea of building a MakerSpace from nothing might sound intimidating, the popularity of the movement actually helps to provide a wide range of technology and affordable options.