César de la Fuente a Presidential Assistant Professor in the Perelman School of Medicine’s departments of Psychiatry and Microbiology and Engineering’s department of Bioengineering, has racked up accolades for his innovative, computational approach to discovering new antibiotics.
Now, in his most recent study, de la Fuente has shown how these vital drugs might be derived from wasp venom.
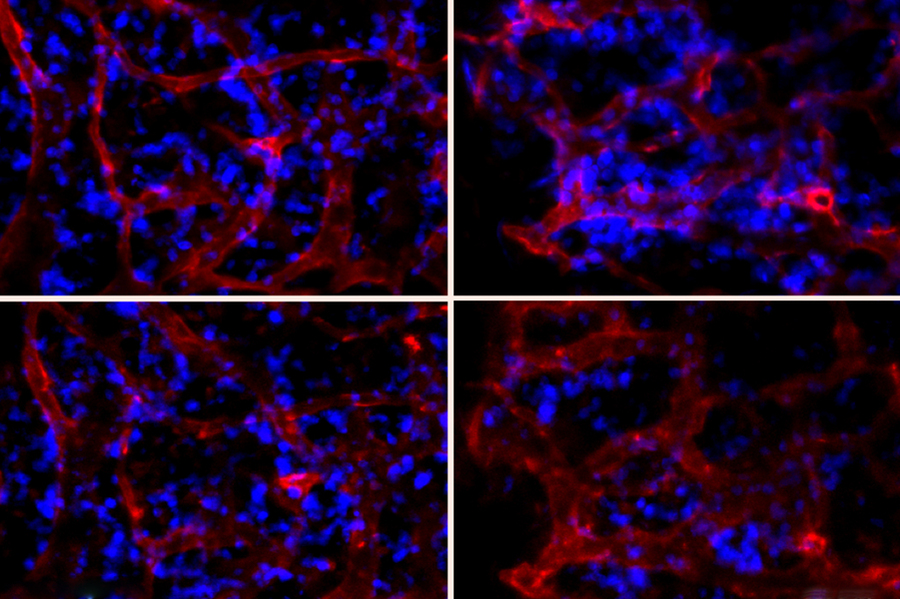
The study, published in The Proceedings of the National Academy of Sciences, involved altering a highly toxic small protein from a common Asian wasp species, Vespula lewisii, the Korean yellow-jacket wasp. The alterations enhanced the molecule’s ability to kill bacterial cells while greatly reducing its ability to harm human cells. In animal models, de la Fuente and his colleagues showed that this family of new antimicrobial molecules made with these alterations could protect mice from otherwise lethal bacterial infections.
There is an urgent need for new drug treatments for bacterial infections, as many circulating bacterial species have developed a resistance to older drugs. The U.S. Centers for Disease Control & Prevention has estimated that each year nearly three million Americans are infected with antibiotic-resistant microbes and more than 35,000 die of them. Globally the problem is even worse: Sepsis, an often-fatal inflammatory syndrome triggered by extensive bacterial infection, is thought to have accounted for about one in five deaths around the world as recently as 2017.
“New antibiotics are urgently needed to treat the ever-increasing number of drug-resistant infections, and venoms are an untapped source of novel potential drugs. We think that venom-derived molecules such as the ones we engineered in this study are going to be a valuable source of new antibiotics,” says de la Fuente.
De la Fuente and his team started with a small protein, or “peptide,” called mastoparan-L, a key ingredient in the venom of Vespula lewisii wasps. Mastoparan-L-containing venom is usually not dangerous to humans in the small doses delivered by wasp stings, but it is quite toxic. It destroys red blood cells, and triggers a type of allergic/inflammatory reaction that in susceptible individuals can lead to a fatal syndrome called anaphylaxis—in which blood pressure drops and breathing becomes difficult or impossible.
Mastoparan-L (mast-L) also is known for its moderate toxicity to bacterial species, making it a potential starting point for engineering new antibiotics. But there are still some unknowns, including how to enhance its anti-bacterial properties, and how to make it safe for humans.
Continue reading at Penn Medicine News.