The paper-based tests could be integrated directly into facemasks and provide instant results at testing sites.

When Penn Health-Tech announced its Nemirovsky Engineering and Medicine Opportunity, or NEMO Prize, in February, the center’s researchers could only begin to imagine the impact the looming COVID-19 pandemic was about to unleash. But with the promise of $80,000 to support early-stage ideas at the intersection of engineering and medicine, the contest quickly sparked a winning innovation aimed at combating the crisis.
Judges from the University of Pennsylvania’s School of Engineering and Applied Sciences and Perelman School of Medicine awarded its first NEMO Prize to César de la Fuente, PhD, who proposed a paper-based COVID diagnostic system that could capture viral particles on a person’s breath, then give a result in a matter of seconds when taken to a testing site.
Similar tests for bacteria cost less than a dollar each to make. De la Fuente, a Presidential Assistant Professor in the departments of Psychiatry, Microbiology, and Bioengineering, is aiming to make COVID tests at a similar price point and with a smaller footprint so that they could be directly integrated into facemasks, providing further incentive for their regular use.
“Wearing a facemask is vital to containing the spread of COVID because, before you know you’re sick, they block your virus-carrying droplets so those droplets can’t infect others,” de la Fuente says. “What we’re proposing could eventually lead to a mask that can be infected by the virus and let you know that you’re infected, too.”
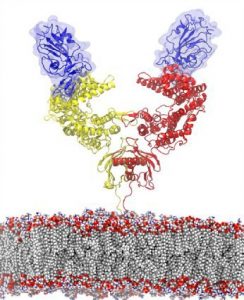
De la Fuente’s expertise is in synthetic biology and molecular-scale simulations of disease-causing viruses and bacteria. Having such fine-grained computational models of these microbes’ binding sites allow de la Fuente to test them against massive libraries of proteins, seeing which bind best. Other machine learning techniques can then further narrow down the minimum molecular structures responsible for binding, resulting in functional protein fragments that are easier to synthesize and manipulate.
The spike-shaped proteins that give coronaviruses their crown-like appearance and name bind to a human receptor known as ACE2. De la Fuente and his colleagues are now aiming to characterize the molecular elements and environmental factors that would allow for the most precise, reliable detection of the virus.
Read the full story on the Penn Engineering blog.
