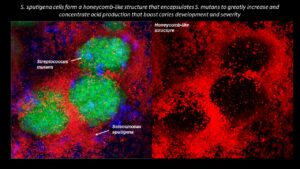
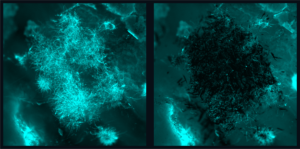
Collaborating researchers from the University of Pennsylvania School of Dental Medicine and the Adams School of Dentistry and Gillings School of Global Public Health at the University of North Carolina have discovered that a bacterial species called Selenomonas sputigena can have a major role in causing tooth decay.
Scientists have long considered another bacterial species, the plaque-forming, acid-making Streptococcus mutans, as the principal cause of tooth decay—also known as dental caries. However, in the study, published in Nature Communications, the Penn Dental Medicine and UNC researchers showed that S. sputigena, previously associated only with gum disease, can work as a key partner of S. mutans, greatly enhancing its cavity-making power.
“This was an unexpected finding that gives us new insights into the development of caries, highlights potential future targets for cavity prevention, and reveals novel mechanisms of bacterial biofilm formation that may be relevant in other clinical contexts,” says study co-senior author Hyun (Michel) Koo, a professor in the Department of Orthodontics and Divisions of Pediatrics and Community Oral Health and co-director of the Center for Innovation & Precision Dentistry at Penn Dental Medicine.
The other two co-senior authors of the study were Kimon Divaris, professor at UNC’s Adams School of Dentistry, and Di Wu, associate professor at the Adams School and at the UNC Gillings School of Global Public Health.
“This was a perfect example of collaborative science that couldn’t have been done without the complementary expertise of many groups and individual investigators and trainees,” Divaris says.
Read the full story in Penn Today.
Michel Koo is a professor in the Department of Orthodontics and divisions of Community Oral Health and Pediatric Dentistry in Penn Dental Medicine and co-director of the Center for Innovation & Precision Dentistry. He is a member of the Penn Bioengineering Graduate Group.



 In
In