by
“This technology is not alive,” says Laia Mogas-Soldevila. “It is living-like.”
The distinction is an important one for the assistant professor at the Stuart Weitzman School of Design, for reasons both scientific and artistic. With a doctorate in biomedical engineering, several degrees in architecture, and a devotion to sustainable design, Mogas-Soldevila brings biology to everyday life, creating materials for a future built halfway between nature and artifice.
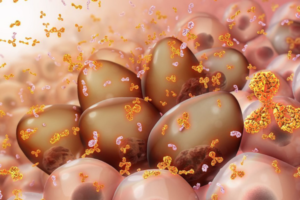
The architectural technology she describes is unassuming at first look: A freeze-dried pellet, small enough to get lost in your pocket. But this tiny lump of matter, the result of more than a year’s collaboration between designers, engineers and biologists, is a biomaterial that contains a “living-like” system.
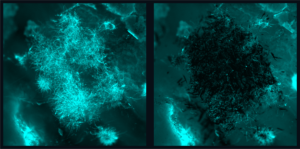
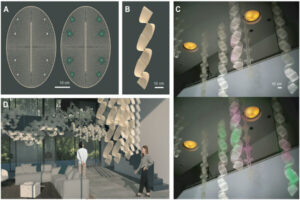
When touched by water, the pellet activates and expresses a glowing protein, its fluorescence demonstrating that life and art can harmonize into a third and very different thing, as ready to please as to protect. Woven into lattices made of flexible natural materials promoting air and moisture flow, the pellets form striking interior design elements that could one day keep us healthy.
“We envision them as sensors,” explains Mogas-Soldevila. “They may detect pathogens, such as bacteria or viruses, or alert people to toxins inside their home. The pellets are designed to interact with air. With development, they could monitor or even clean it.”
For now, they glow, a triumphant first stop on the team’s roadmap to the future. The fluorescence establishes that the lab’s biomaterial manufacturing process is compatible with the leading-edge cell-free engineering that gives the pellets their life-like properties.
A rapidly expanding technology, cell-free protein expression systems allow researchers to manufacture proteins without the use of living cells.
Gabrielle Ho, Ph.D. candidate in the Department of Bioengineering and co-leader of the project, explains how the team’s design work came to be cell-free, a technique rarely explored outside of lab study or medical applications.
“Typically, we’d use living E. coli cells to make a protein,” says Ho. “E. coli is a biological workhorse, accessible and very productive. We’d introduce DNA to the cell to encourage expression of specific proteins. But this traditional method was not an option for this project. You can’t have engineered E. coli hanging on your walls.”
Cell-free systems contain all the components a living cell requires to manufacture protein —energy, enzymes and amino acids — and not much else. These systems are therefore not alive. They do not replicate, and neither can they cause infection. They are “living-like,” designed to take in DNA and push out protein in ways that previously were only possible using living cells.
“One of the nicest things about these materials not being alive,” says Mogas-Soldevila, “is that we don’t need to worry about keeping them that way.”
Unlike living cells, cell-free materials don’t need a wet environment or constant monitoring in a lab. The team’s research has established a process for making these dry pellets that preserves bioactivity throughout manufacturing, storage and use.
Bioactive, expressive and programmable, this technology is designed to capitalize on the unique properties of organic materials.
Mogas-Soldevila, whose lab focuses exclusively on biodegradable architecture, understands the value of biomaterials as both environmentally responsible and aesthetically rich.
“Architects are coming to the realization that conventional materials — concrete, steel, glass, ceramic, etc. — are environmentally damaging and they are becoming more and more interested in alternatives to replace at least some of them. Because we use so much, even being able to replace a small percentage would result in a significant reduction in waste and pollution.”
Her lab’s signature materials — biopolymers made from shrimp shells, wood pulp, sand and soil, silk cocoons, and algae gums — lend qualities over and above their sustainable advantages.
“My obsession is diagnostic, but my passion is playfulness,” says Mogas-Soldevila. “Biomaterials are the only materials that can encapsulate this double function observed in nature.”
This multivalent approach benefited from the help of Penn Engineering’s George H. Stephenson Foundation Educational Laboratory & Bio-MakerSpace, and the support of its director, Sevile Mannickarottu. In addition to contributing essential equipment and research infrastructure to the team, Mannickarottu was instrumental in enabling the interdisciplinary relationships that led the team to success, introducing Ho to the DumoLab Research team collaborators. These include Mogas-Soldevila, Camila Irabien, a Penn Biology major who provided crucial contributions to experimental work, and Fulbright design fellow Vlasta Kubušová, who co-led the project during her time at Penn and who will continue fueling the project’s next steps.