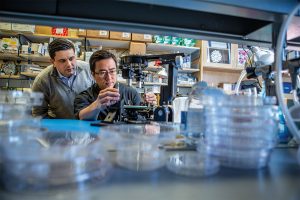
With Vivodyne, Associate Professor in the Department of Bioengineering Dan Huh is translating the organs-on-chips technology into a promising industry venture. Using microfluidic structures that mimic aspects of human physiology, organs-on-chips allow scientists to test therapies on lab-grown human cells. Vivodyne specifically focuses on designing organs-on-chips to create a scalable alternative for pharmaceutical drug testing on animals.
Last year, the company raised $4 million dollars in seed money. This year, it’s topping influential lists of small companies making big impacts.
Fast Company now lists it as one of “the 10 most innovative companies with fewer than 10 employees,” saying “Vivodyne is helping major pharmaceutical companies like GlaxoSmithKline quickly adopt viable alternatives for testing drugs on monkeys.”
Vivodyne, launched in 2021, has created a platform that allows fully automated, complex studies at a far larger scale and lower cost than would be possible with manual experimentation, so pharmaceutical companies can actually test lab-made organs instead of animals in their drug-development processes. When done by hand, only 20 to 40 living tissue samples can be managed in parallel; Vivodyne’s instrument can cultivate, dose, and image more than 2,000 living tissues at once. The company, which raised $4 million in seed funding last year, says its instruments currently play pivotal roles in clinical drug testing for respiratory diseases, cancer treatment, vaccine development, diabetes therapies, and maternal medicine. GlaxoSmithKline, one of Vivodyne’s clients, estimates that for some projects the lab-grown tissues may displace as much as 80% of its animal testing. The company’s ultimate goal? “To supplant the vast majority of animal testing within the next decade,” says CEO Andrei Georgescu.
Continue reading “The 10 most innovative companies with fewer than 10 employees” at Fast Company.
Originally posted in Penn Engineering today.





 NextUp, a regular feature of Philadelphia Magazine that “highlights the local leaders, organizations and research shaping the Greater Philadelphia region’s life sciences ecosystem,” ran a profile of Philly-based agricultural startup Strella Biotechnology. Founded by Penn alumna Katherine Sizov (Bio 2019) and winner of a 2019 President’s Innovation Prize, Strella Biotech seeks to reduce food waste through innovative biosensors, and was initially developed in the
NextUp, a regular feature of Philadelphia Magazine that “highlights the local leaders, organizations and research shaping the Greater Philadelphia region’s life sciences ecosystem,” ran a profile of Philly-based agricultural startup Strella Biotechnology. Founded by Penn alumna Katherine Sizov (Bio 2019) and winner of a 2019 President’s Innovation Prize, Strella Biotech seeks to reduce food waste through innovative biosensors, and was initially developed in the