 by Beth Adams
by Beth Adams
With the shared vision to transform the future of oral health care, Penn Dental Medicine and Penn’s School of Engineering and Applied Sciences have united to form the Center for Innovation & Precision Dentistry (CiPD). The new Center marked its official launch on January 22 with a virtual program celebrating the goals and plans of this unique partnership. Along with the Deans from both schools, the event gathered partners from throughout the University of Pennsylvania and invited guests, including the National Institute of Dental and Craniofacial Research Director (NIDCR) Dr. Rena D’Souza and IADR Executive Director Chris Fox.
Conceived and brought to fruition by co-directors Dr. Michel Koo of Penn Dental Medicine and Dr. Kathleen Stebe of Penn Engineering, the CiPD is bridging the two schools through cutting-edge research and technologies to accelerate the development of new solutions and devices to address unmet needs in oral health, particularly in the areas of dental caries, periodontal disease, and head and neck cancer. The CiPD will also place a high priority on programs to train the next generation of leaders in oral health care innovation.
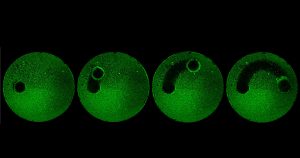
 “We have a tremendous global health challenge. Oral diseases and craniofacial disorders affect 3.5 billion people, disproportionately affecting the poor and the medically and physically compromised,” says Dr. Koo, Professor in the Department of Orthodontics and Divisions of Community Oral Health and Pediatric Dentistry, in describing their motivation to form the Center. “There is an urgent need to find better ways to diagnose, prevent, and treat these conditions, particularly in ways that are affordable and accessible for the most susceptible populations. That is our driving force for putting this Center together.”
“We have a tremendous global health challenge. Oral diseases and craniofacial disorders affect 3.5 billion people, disproportionately affecting the poor and the medically and physically compromised,” says Dr. Koo, Professor in the Department of Orthodontics and Divisions of Community Oral Health and Pediatric Dentistry, in describing their motivation to form the Center. “There is an urgent need to find better ways to diagnose, prevent, and treat these conditions, particularly in ways that are affordable and accessible for the most susceptible populations. That is our driving force for putting this Center together.”
“We have united our schools around this mission,” adds Dr. Stebe, Richer & Elizabeth Goodwin Professor in the Department of Chemical and Biomolecular Engineering. “We have formed a community of scholars to develop and harness new engineering paradigms, to generate new knowledge, and to seek new approaches that are more effective, precise, and affordable to address oral health. More importantly, we will train a new community of scholars to impact this space.”
Born through Interdisciplinary Research
A serendipitous connection born through Penn’s interdisciplinary research environment itself brought Drs. Koo and Stebe together more than five years ago, an introduction that would eventually lead to creating the CiPD.
Dr. Tagbo Niepa, now assistant professor at the University of Pittsburgh, came to Penn Engineering in 2014 as part of Penn’s Postdoctoral Fellowship for Academic Diversity, an initiative from the office of the Vice Provost for Research. His studies on the microbiome led him to reach out to Dr. Stebe and Dr. Daeyeon Lee (also at Penn Engineering), and to connect them to Dr. Koo, initiating collaboration between their labs.
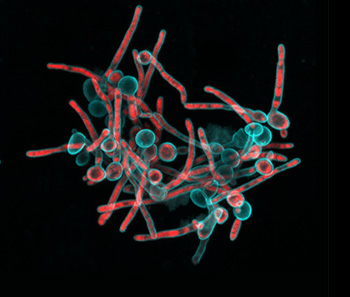 “Tagbo embodies what we are trying to do with the CiPD,” recalls Dr. Stebe. “He had initiative, he identified new tools and important context, and he did good science that may help us understand how to interrupt the disease process and identify new underlying mechanisms that can inspire new therapies.” Dr. Niepa worked on applying microfluidics and engineering to study the oral microbiome and better understand how the interactions between fungi and bacteria could impact dental caries.
“Tagbo embodies what we are trying to do with the CiPD,” recalls Dr. Stebe. “He had initiative, he identified new tools and important context, and he did good science that may help us understand how to interrupt the disease process and identify new underlying mechanisms that can inspire new therapies.” Dr. Niepa worked on applying microfluidics and engineering to study the oral microbiome and better understand how the interactions between fungi and bacteria could impact dental caries.
“Upon meeting Michel, we became excited about the possibilities of bringing talent from the two schools together,” notes Dr. Stebe. A 2018 workshop organized by Drs. Koo and Stebe and funded by Penn’s Vice Provost of Research explored the potential for expanding cross-school research. “We invited researchers from dental medicine and engineering as well as relevant people from the arts and sciences to see if we could find a way to collaborate to advance oral and craniofacial health,” says Dr. Koo. “That was the catalyst for the Center; after the workshop, we put together a task force which would become the core members of the CiPD.”
In addition to Drs. Koo and Stebe, the CiPD Executive Committee includes Associate Directors Dr. Henry Daniell, Vice-Chair and W.D. Miller Professor, Department of Basic & Translational Sciences, Penn Dental Medicine, and Dr. Anh Le, Chair and Norman Vine Endowed Professor of Oral Rehabilitation, Department of Oral and Maxillofacial Surgery / Pharmacology, Penn Dental Medicine; as well as Dr. Andrew Tsourkas, Professor, Department of Bioengineering, Co-Director, Center for Targeted Therapeutics & Translational Nanomedicine (CT3N) and Chemical and Nanoparticle Synthesis Core, Penn Engineering; and Dr. Jason Moore, Edward Rose Professor of Informatics, Director of the Penn Institute for Biomedical Informatics. The core members of CiPD include 26 faculty from across both Penn Dental Medicine and Penn Engineering, and also from the Schools of Medicine and Arts & Sciences.
Read the full story in Penn Today.


 New research from
New research from 





 by Beth Adams
by Beth Adams “We have a tremendous global health challenge. Oral diseases and craniofacial disorders affect 3.5 billion people, disproportionately affecting the poor and the medically and physically compromised,” says Dr. Koo, Professor in the Department of Orthodontics and Divisions of Community Oral Health and Pediatric Dentistry, in describing their motivation to form the Center. “There is an urgent need to find better ways to diagnose, prevent, and treat these conditions, particularly in ways that are affordable and accessible for the most susceptible populations. That is our driving force for putting this Center together.”
“We have a tremendous global health challenge. Oral diseases and craniofacial disorders affect 3.5 billion people, disproportionately affecting the poor and the medically and physically compromised,” says Dr. Koo, Professor in the Department of Orthodontics and Divisions of Community Oral Health and Pediatric Dentistry, in describing their motivation to form the Center. “There is an urgent need to find better ways to diagnose, prevent, and treat these conditions, particularly in ways that are affordable and accessible for the most susceptible populations. That is our driving force for putting this Center together.” “Tagbo embodies what we are trying to do with the CiPD,” recalls Dr. Stebe. “He had initiative, he identified new tools and important context, and he did good science that may help us understand how to interrupt the disease process and identify new underlying mechanisms that can inspire new therapies.” Dr. Niepa worked on applying microfluidics and engineering to study the oral microbiome and better understand how the interactions between fungi and bacteria could impact dental caries.
“Tagbo embodies what we are trying to do with the CiPD,” recalls Dr. Stebe. “He had initiative, he identified new tools and important context, and he did good science that may help us understand how to interrupt the disease process and identify new underlying mechanisms that can inspire new therapies.” Dr. Niepa worked on applying microfluidics and engineering to study the oral microbiome and better understand how the interactions between fungi and bacteria could impact dental caries.