Imagine the brain as an air traffic control tower, overseeing the crucial and complex operations of the body’s ‘airport.’ This tower, essential for coordinating the ceaseless flow of neurological signals, is guarded by a formidable layer that functions like the airport’s security team, diligently screening everything and everyone, ensuring no unwanted intruders disrupt the vital workings inside.
However, this security, while vital, comes with a significant drawback: sometimes, a ‘mechanic’—in the form of critical medication needed for treating neurological disorders—is needed inside the control tower to fix arising issues. But if the security is too stringent, denying even these essential agents entry, the very operations they’re meant to protect could be jeopardized.
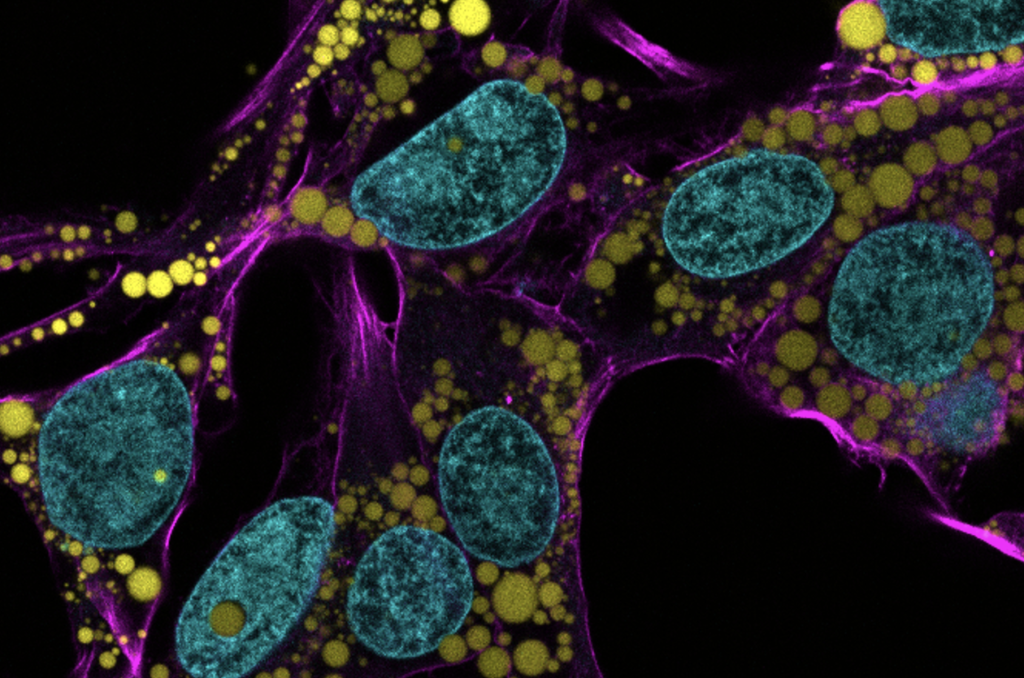
Now, researchers led by Michael Mitchell of the University of Pennsylvania are broaching this long-standing boundary in biology, known as the blood-brain barrier, by developing a method akin to providing this mechanic with a special keycard to bypass security. Their findings, published in the journal Nano Letters, present a model that uses lipid nanoparticles (LNPs) to deliver mRNA, offering new hope for treating conditions like Alzheimer’s disease and seizures—not unlike fixing the control tower’s glitches without compromising its security.
“Our model performed better at crossing the blood-brain barrier than others and helped us identify organ-specific particles that we later validated in future models,” says Mitchell, associate professor of bioengineering at Penn’s School of Engineering and Applied Science, and senior author on the study. “It’s an exciting proof of concept that will no doubt inform novel approaches to treating conditions like traumatic brain injury, stroke, and Alzheimer’s.”
Read the full story in Penn Today.